
enriched with
FGFR3 mutations,
amplification,
and
expres-
sion
and
urothelial
differentiation
markers
(UPK1A,2,3A
and
KRT20).
Cluster
III
is
enriched
with
squamous
morphology,
basal
markers
(KRT
14,
KRT
5,
TP63),
and
expression of several
immune
response genes. We have also
performed unsupervised clustering of miRNA
in 310
tumors
and
provide
an
integrated
analysis
of
anticorrelation
of
clusters
of
miRNA
regulating
many
pathways
including
epithelial–mesenchymal
transition, DNA methylation,
and
FGFR3
expression,
among many
others.
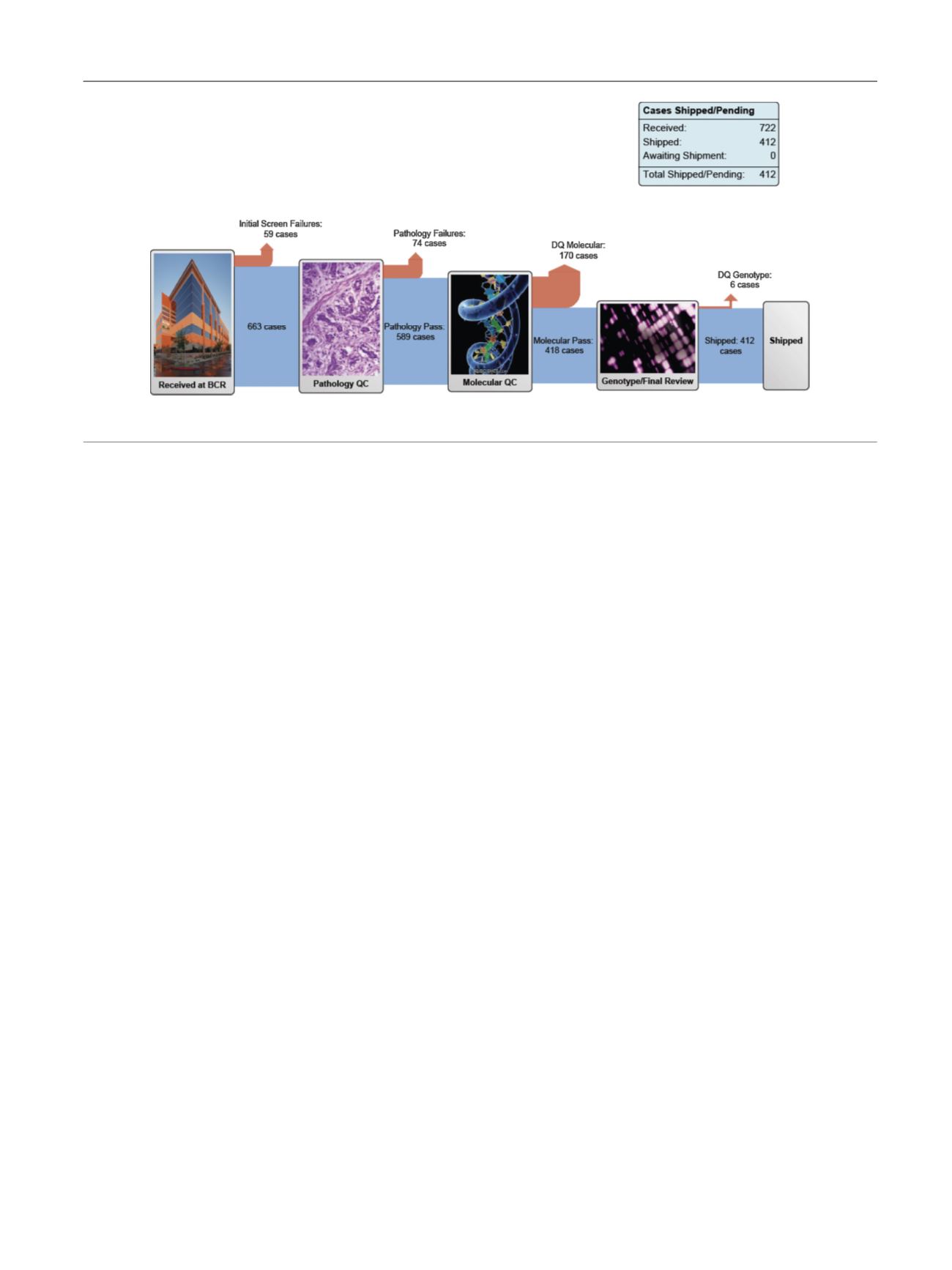
The
total
cohort now
includes 412
tumors
that have met
the
prespecified
quality
controls
for
pathology
and
RNA
quality
and
have
been
distributed
to
the
genome
charac-
terization
centers
[6] ( Fig. 1).
The
complete
data
set will
increase
the
power
to
detect
additional
low-frequency
events
[7],
validate
the
cluster
analyses,
test
hypotheses
regarding
chemotherapy
resistance,
and
provide
a
host
of
translational
opportunities
for
functional
validation
and
targeted
therapy
trials. Outcome analyses were deliberately
not
included
in
the
analysis
of
the
first
131
tumors,
as
the
follow-up
data were
not mature. We
expect
to
be
able
to
include
this
in
the
final
analysis
of
the
full
cohort.
The
analysis working
group
reconvened
in
early
2015
to
begin
the
final
analysis,
with
the
expectation
of
publishing
an
updated
comprehensive
integrated
analysis.
There
is
a
large unmet need
for
comprehensive genomic
characterization
of
non–muscle-invasive
bladder
cancer.
FGFR3
mutations
characterize
low-grade
Ta
tumors,
and
high-grade
tumors
share
similar
genomic
alterations with
muscle-invasive
carcinomas.
The National
Cancer
Institute
sponsored
a
clinical
trial
planning meeting
held
in March,
2015, with
one
of
the
goals
to
design
a
targeted
therapy
trial. A more
comprehensive understanding of
the genomic
landscape
is a critical step
in
this process. A recent
landmark
study
of
aristolochic
acid–induced
upper
tract
tumors,
using whole-exome
sequencing,
found
a
remarkably
high
somatic
mutation
rate
and
a
unique
mutation
signature
[8].
There
are
funding
opportunities
specifically
for
rare
cancers
(3–6
per
100
000),
and
this
presents
unique
collaborative
opportunities
for
important
research
on
the
biology
and
treatment
of
upper
tract
urothelial
carcinoma
[9].
Conflicts
of
interest:
The
author
has
nothing
to
disclose.
References
[1]
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507: 315–22.
[2]
Volkmer JP, Sahoo D, Chin RK, et al. Three differentiation states risk- stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci U S A 2012;109:2078–83.[3]
HoPL,Kurtova A, ChanKS. Normal andneoplastic urothelial stemcells: getting to the root of the problem. Nat Rev Urol 2012;9:583–94.
[4]
Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012;18:3377–86.
[5]
Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 can- cer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929–44.
[6]
BCR
pipeline
report.
The
Cancer
Genome
Atlas Web
site.
https:// tcga-data.nci.nih.gov/datareports/BCRPipelineReport.htm .Accessed
November
8,
2014.
[7]
Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505: 495–501.[8]
Hoang ML, Chen CH, Sidorenko VS, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med 2013;5:197ra02.[9]
Participation
in Trials of Rare Cancers. National Cancer
Institute Web
site.
http://cancercenters.cancer.gov/news/news-announ-comm. html .Fig.
1
–
The
Cancer
Genome Atlas muscle-invasive
bladder
cancer
pipeline. Reproduced with
permission
from
the National
Cancer
Institute
[6].
E U R O P E A N
U R O L O G Y
F O C U S
1
( 2 0 1 5
)
9 4 – 9 5
95

















